Computer Graphics Forum (Proc. EuroVis), 2018
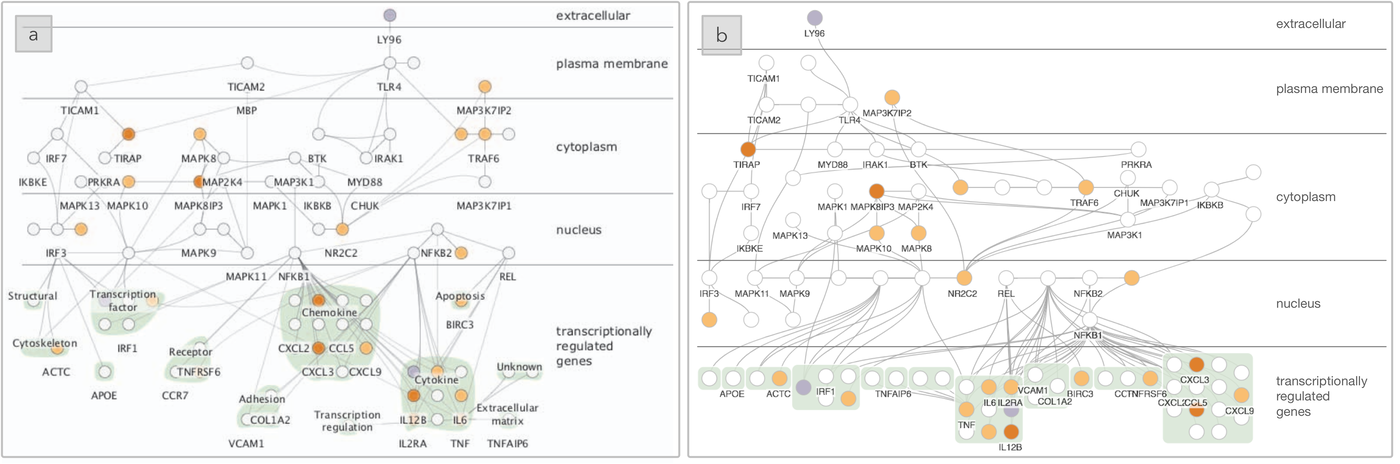
The layout for the TLR4 biological system produced using (a) Cerebral, a domain-specific layout tool, as compared to (b) SetCoLa. The layers correspond to the location of the biomolecule within a cell and show immune response outcomes at the bottom of the graph, grouped by molecular function.
Abstract
Constraints enable flexible graph layout by combining the ease of automatic layout with customizations for a particular domain. However, constraint-based layout often requires many individual constraints defined over specific nodes and node pairs. In addition to the effort of writing and maintaining a large number of similar constraints, such constraints are specific to the particular graph and thus cannot generalize to other graphs in the same domain. To facilitate the specification of customized and generalizable constraint layouts, we contribute SetCoLa: a domain-specific language for specifying high-level constraints relative to properties of the backing data. Users identify node sets based on data or graph properties and apply high-level constraints within each set. Applying constraints to node sets rather than individual nodes reduces specification effort and facilitates reapplication of customized layouts across distinct graphs. We demonstrate the conciseness, generalizability, and expressiveness of SetCoLa on a series of real-world examples from ecological networks, biological systems, and social networks.